McKenna Feltes
Postdoctoral Associate
Johns Hopkins University
About Me
McKenna Feltes is a postdoctoral associate in the laboratory of Dr. Steven Farber at Johns Hopkins University. Her research focuses on understanding the cellular machinery responsible for building and regulating lipoproteins, fat-containing particles that contribute to human disease. She uses the zebrafish model and a variety of cell biological and physiological techniques to identify and investigate genes with previously unappreciated roles in lipoprotein biosynthesis.
Interests
- lipoprotein biology
- secretory pathway
- genetics
Education
-
PhD Biochemistry, Biophysics & Structural Biology
Washington University
-
BA, summa cum laude, Biology & Chemistry
Drury University
My Research
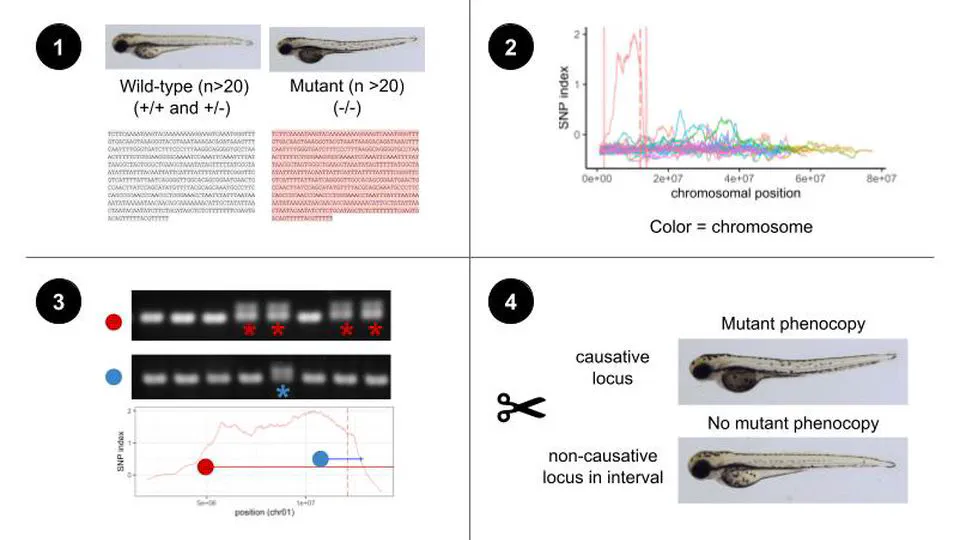
I’m a postdoctoral associate at Johns Hopkins University in the laboratory of Dr. Steven Farber, where I seek to identify and characterize new genes involved in lipoprotein biogenesis. Zebrafish are a powerful model for the study of lipoprotein biology - lipoprotein machinary is conserved between zebrafish and humans, and the optical transparency of the larval zebrafish allows us to observe cell-level biology in a whole organism. In larval stages, zebrafish produce lipoproteins from maternally deposited yolk lipid in a tissue called the yolk syncitial layer. The yolk syncitial layer serves as the zebrafish’s first digestive organ, and expresses many genes that are later turned on in the liver and intestine once this tissues have developed. When lipoprotein synthesis is disrputed (i.e.: by mutation or drug inhibition), yolk lipid is no longer processed into lipoproteins and accumulates in the yolk syncitial layer instead, changing its optical properties. We leveraged this so-called “dark yolk” phenotype and the power of foward genetics to identify 28 novel dark yolk alleles. The mutant collection includes 10 alleles at known dark yolk loci (mttp, apobb.1, dgat2, mia2) as well as at least 13 additional novel loci. We developed a rapid mutation mapping pipieline called WheresWalker which we have so far used to identify 4 of the unknown loci. Using a suite of optical rerporters, genetic and small molecule manipulations, and human GWAS datasets, we are begnniing to uncover the mechanism by which these novel dark yolk genes contribute to lipoprotein metabolism.
Featured Publications
Recent Publications
(2021).
Validation of Trifluoromethylphenyl Diazirine Cholesterol Analogues As Cholesterol Mimetics and Photolabeling Reagents.
ACS chemical biology.
(2020).
Monitoring the itinerary of lysosomal cholesterol in Niemann-Pick Type C1-deficient cells after cyclodextrin treatment [S].
Journal of lipid research.
(2019).
Synthesis and characterization of diazirine alkyne probes for the study of intracellular cholesterol trafficking [S].
Journal of lipid research.
(2018).
High-content screen for modifiers of Niemann-Pick type C disease in patient cells.
Human molecular genetics.